Working with Cancer Genomics Cloud datasets in a PostgreSQL database (Part 1)
Posted on Mon 12 October 2020 in SQL
Introduction
Recently I have been looking for publicly-available genomics datasets to test machine learning models in Python. During my searches for such a “toy dataset”, I came upon the Cancer Genomics Cloud (CGC) initiative.
Anyone can register in CGC and have access to open access massive public datasets, like The Cancer Genomics Atlas (TCGA). Most individual-level genomic data can only be accessed following approval of a Data Access Request through the Database of Genotypes and Phenotypes (dbGaP). For now, I guess the open data tier will suffice for this exercise.
This demonstration will be separated into two parts. Here in the first part I will provide a brief run-down of how I queried the CGC to obtain genomic data from cancer patients and the first steps into preparing a local PostgreSQL relational database in my computer.
Querying the CGC
I registered at CGC, then I created a project in the CGC dashboard and went into the data browser tool:
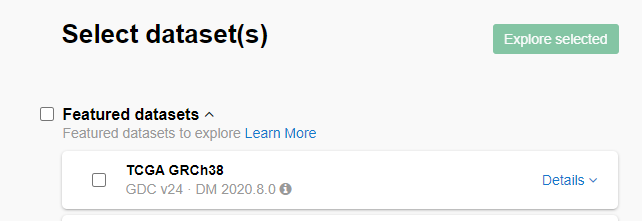
Then, I chose the TCGA GRCh38 dataset and clicked on the Explore selected button.
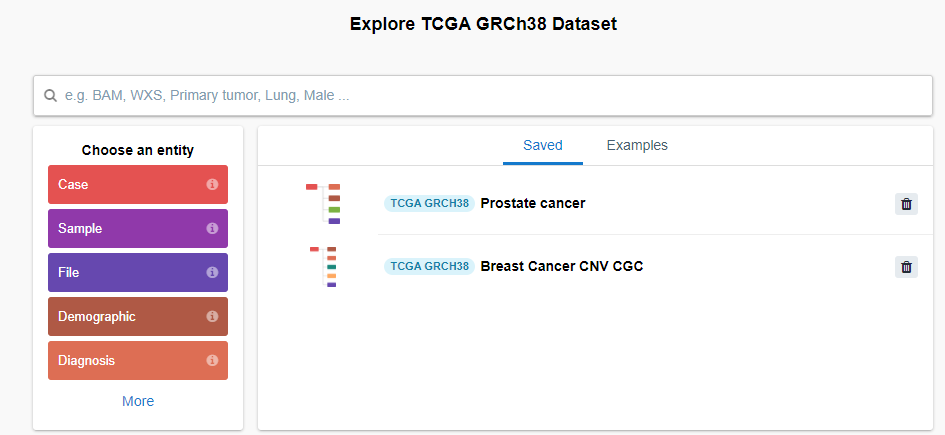
Inside the data browser, I see that there are several information entities:
I clicked on the first one, Cases and then created a query with the following entities and filters:
- Entity
- Filters
- Case
- Primary site: Prostate Gland
- Diagnosis
- Age at diagnosis
- Clinical T (TNM)
- Demographic
- Ethnicity
- Race
- Follow up
- Primary therapy outcome
- File
- Access level: Open
- Data type: Gene Level Copy Number, Gene Expression Quantification, Gene Level Copy Number Scores
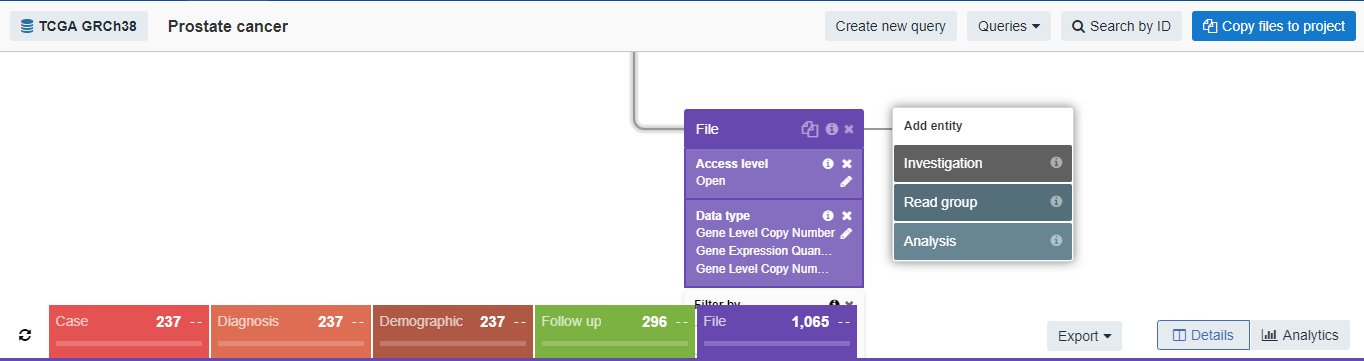
The final query ended up like this:
In other words, the query resulted in individuals diagnosed with prostate cancer (n=237), their age at diagnosis, their demographic characteristics, their therapeutic outcomes, and their genomic data (n=1,065 files overall: 276 with raw counts of gene expression quantification, 552 with FPKM information, 236 from copy number variation genotyping, and a single file containing what I believe is a prostate cancer diagnosis score stratified by gene).
Then, I clicked on the Copy files to project and on the Export button and chose Export as TSV option. I went back to my project dashboard, clicked on the Files tab and downloaded everything.
I realized that the four TSV and the genomic data could be organized as tables on a relational database. So I used my PostgreSQL server that I have installed on computer. For this demonstration, I will use my Windows 10 OS, but PostgreSQL can be installed on Unix systems as well. In my portfolio I provide a Windows script and a Unix script as well containing the steps I followed to load all the data into a PostgreSQL database.
Creating the ‘tcga’ database into the local PostgreSQL server
The official PostgreSQL installation instructions are here.
I created a folder named TCGA for this project, and put the downloaded files inside a data subfolder. Here is a representation of my directory structure:
.
└── TCGA
├── data
│ ├── counts
│ │ └── [200+ *.counts.gz files]
│ ├── focal_score_by_genes
│ ├── fpkm
│ ├── gene_level_copy_numbers
│ ├── cases.tsv
│ ├── demographic.tsv
│ ├── files.tsv
│ └── follow_up.tsv
├── main_tcga.ps1
└── main_tcga.sh
The TSV files are the query results and inside the folders are the files containing the genomic data (these are not their original names — I renamed them to make easier to identify the contents of each one). For now, I will use just the four TSV files and the counts folders.
This structure is replicated in the folder corresponding to this post in my portfolio. The main_tcga.ps1 and main_tcga.sh files contain the commands I used for this demonstration. The first is for Windows and the second for Unix systems.
Then, in the TCGA folder I opened a Windows PowerShell terminal and using psql, a terminal-based front-end to PostgreSQL, created a database named tcga on my local server:
psql -U postgres -c "CREATE DATABASE tcga ENCODING 'UTF-8' LC_COLLATE 'English_United States' LC_CTYPE 'English_United States' TEMPLATE template0"
The -U flag serves to indicate which user will connect to the local PostgreSQL server. postgres is the default user created during PostgreSQL installation. The -c flag means that we are sending a command to the server. Note that the command is inside double quotes and strings into the command are single-quoted.In summary, this command serves to connect the postgres user into the server and pass a command to create the tcga database with certain characteristics: use UTF-8 codification, with English locale using the template0 database as template, which is created by default during PostgreSQL server installation.
If during installation you provided a password to access the server, the terminal will ask for it after you press Enter.
Creating tables in the ‘tcga’ database
Then, I created four tables, corresponding to each TSV files with the following command:
psql -U postgres -d tcga -a -f "src/tcga_create_tables.sql"
The new friends here are -d and -a -f. -d is the flag that indicates the database I wished to connect; it is the tcga I created above. The -a serves to echo all information from the command to the terminal output so it is possible to check if the commands worked. The -f flag mean file: I am indicating that I want to pass the commands within the tcga_create_tables.sql file inside the src directory — which I created as a subfolder of the TCGA folder. If you are wondering how I created this file: wrote the commands in a text file and simply saved it with the .sql extension.
Below is one of the commands inside the .sql file:
CREATE TABLE allcases (
#case_id TEXT,
#case_primarysite TEXT,
#diagnosis TEXT,
#diagnosis_ageatdiagnosis_1 INT,
#diagnosis_clinicalt_1 TEXT
);
The command above creates the table allcases with five columns: case_id, case_primarysite, diagnosis, diagnosis_ageatdiagnosis_1, and diagnosis_clinicalt_1. Notice the words beside each one: they indicate the data type of the data that the column will hold. In this case I have four columns that will get text data (TEXT) and one that will get numbers — integers (INT) specifically.
Note the semicolon ; at the end — it is a PostgreSQL requirement. It indicates the end of a command (however, if we are passing arguments through the -c flag the semicolon is not needed though, it is implicit within the flag).
The file have three more commands similar to the one above. The output of the second to last command should be CREATE TABLE messages, meaning all went well — I created tour tables inside the tcga database.
Populating the tables
However, they are still empty. To populate the tables, I used the four commands below, one for each table (allcases, demographic, follow_up and allfiles):
psql -U postgres -d tcga -c "\COPY allcases FROM 'data/cases.tsv' DELIMITER E'\t' CSV HEADER"
psql -U postgres -d tcga -c "\COPY demographic FROM 'data/demographic.tsv' DELIMITER E'\t' CSV HEADER"
psql -U postgres -d tcga -c "\COPY follow_up FROM 'data/follow_up.tsv' DELIMITER E'\t' CSV HEADER"
psql -U postgres -d tcga -c "\COPY allfiles FROM 'data/files.tsv' DELIMITER E'\t' CSV HEADER"
(It is good practice to separate table-creating commands of table-populating ones). In summary, the commands tell the PostgreSQL server to copy the information contained in the TSV files inside the data directory into the specified table.
The argument DELIMITER E'\t' means that the columns are tab-separated (delimited). This argument would be DELIMITER ',' if the file were comma-separated or omitted altogether.
The CSV indicates that we are importing a delimiter-separated file. HEADER means that the copied file have a header — the first line have the column titles, which must be equal to the ones specified during table creation; an error will occur otherwise. This argument must be omitted if the file does not have a header.
The output COPY followed by an integer (representing the number of rows copied) means that everything went well. Be careful: do not run the copy commands more than once, otherwise data duplication will occur.
With this I conclude the first part of this demonstration. In the next part I will use I will use a customized Python to help with the import of genomic data into the PostgreSQL database.
Conclusion of Part 1
In this part I:
- Demonstrated how to query open access data in CGC;
- Showed basic commands for importing data into tables created in a local PostgreSQL database.
References
The Cancer Genome Atlas Program
RPKM, FPKM and TPM, clearly explained
Copy Number Variation | Scitable by Nature Education
Relational database - Wikipedia